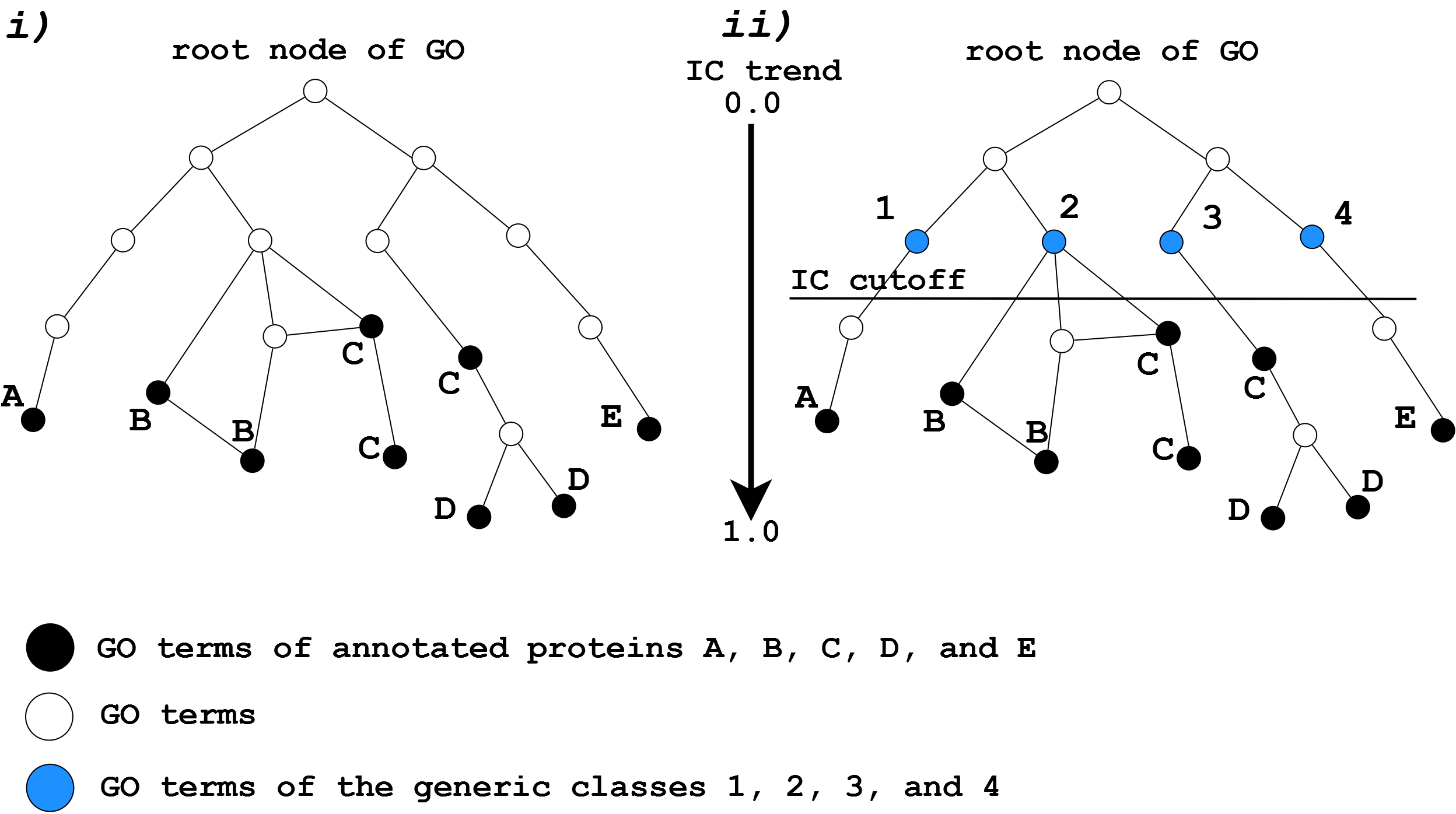
GOClass
GOClass works on simple classification of GO terms based on their Information Content (IC) (see definition here). The aim is to find generic functional classes that can group proteins according to their shared annotations. Each GO term belonging to the annotated protein is traced back up to the root node. During the path reconstruction, a parent node will be considered as the generic “class node” (see azure circle in Fig 1-ii) for that GO term if it has an IC lower than the chosen cutoff. In this way, a sort of GO terms grouping/clustering occurs. As shown in Fig. 1 “class nodes” 1, 2, 3, and 4 subsume and consequently cluster the GO terms belonging to proteins A, B+C, D+C, and E respectively. Due to the distribution of annotations and graph connections, GO terms belonging to the same protein can be part of different classes as shown for protein C whose nodes belong to both class 2 and 3 (see Fig. 1). In this case, the protein is inevitably numbered both in class 2 and class 3.
In the path reconstruction the user can select only those GO predictions having at least a certain TS score (the default is set to 2.0) and can choose a certain IC cutoff. The higher the IC cutoff (up to 1.0) the more classes will be returned whereas the lower the IC cutoff (up to 0.0 corresponding to the root node and consequently the grouping into the “root” class) the more general classes will be retrieved. We suggest an IC of 0.1 as default.