Argot2 Assessment
Different scoring strategies have been used to evaluate Argot2 performance. We partially followed the “Critical Assessment of Function Annotations” (CAFA) experiment guidelines (http://biofunctionprediction.org/) and tested over 4000 proteins, with already available GO annotations in GOA, both from Eukaryota and Prokaryota randomly picked up from about 50000 sequences released for the CAFA challenge. In addition, the well annotated yeast genome, comprising 6187 annotated proteins, has also been used as test set.
The evaluation has been carried out at protein-centric level using the following criteria.
Let N be a pool of unknown target proteins. For each given protein i, the GO terms j predicted by each method are retrieved and ranked accordingly to the corresponding scores tij, 0≤tij≤tmax where tmax is the max score obtained by each method.
For a given score t, the different methods are assessed based on precision and recall, calculated for each protein i as:
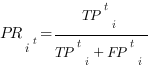 | 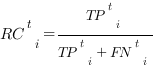 | (1) |
where all the terms j with score tij ≥ t are considered. The number of True Positives (TPti) is the size of the intersection between the sets of predicted and benchmark (true) GO terms; the number of False Positives (FPti) is the size of the difference between the sets of predicted and true GO terms; the number of False Negatives (FNti) is the size of the difference between the sets of true and predicted GO terms. Note that if, for a given t, a protein has not any annotated term, its precision cannot be defined and, therefore, it will not contribute to the averaged precision (see below Eq. 2). The denominators of Eqs. 1 represent the total number of predicted terms and the number of true terms, respectively.
Method 1 (m1) with sliding threshold
We considered a set of threshold values t ranging from 0 to the maximum score obtained by each method. For a given value of t, precision and recall are averaged across the N proteins of the pool, obtaining:
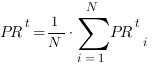 | 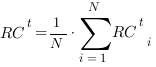 | (2) |
The procedure is repeated for each value t of the grid. Each pair of values (1-PRt,RCt) represents a point of the precision/recall curve.
Method 2 (m2) with sliding threshold
We calculated precision and recall in the case of “m1” method, but predicted and true GO terms are first propagated to the root.
Method 3 (m3) with first k hits
Even in this case, predicted and benchmark GO terms are propagated to the root but, instead of considering for each protein i a set of threshold values t ranging from 0 to tmax, the first k ranked GO terms of each protein, with k=1,2,...,100, were used to calculate precision and recall. Each pair of values (1-PRk,RCk) represents a point of the precision/recall curve.
Compared to “m2” and “m3”, “m1” favors methods that annotate with specific GO terms. As opposed to “m3”, “m1” and “m2” take account for the scoring metrics. Methods “m2” and “m3” consider inexact and shallow predictions that are in parent-child relationship with the original nodes, and consequently would not be completely wrong.
Precision and recall are calculated for each protein in the three ontologies, Molecular Function (MF), Biological Process (BP) and Cellular Component (CC).