FunTaxIS-lite working pipeline
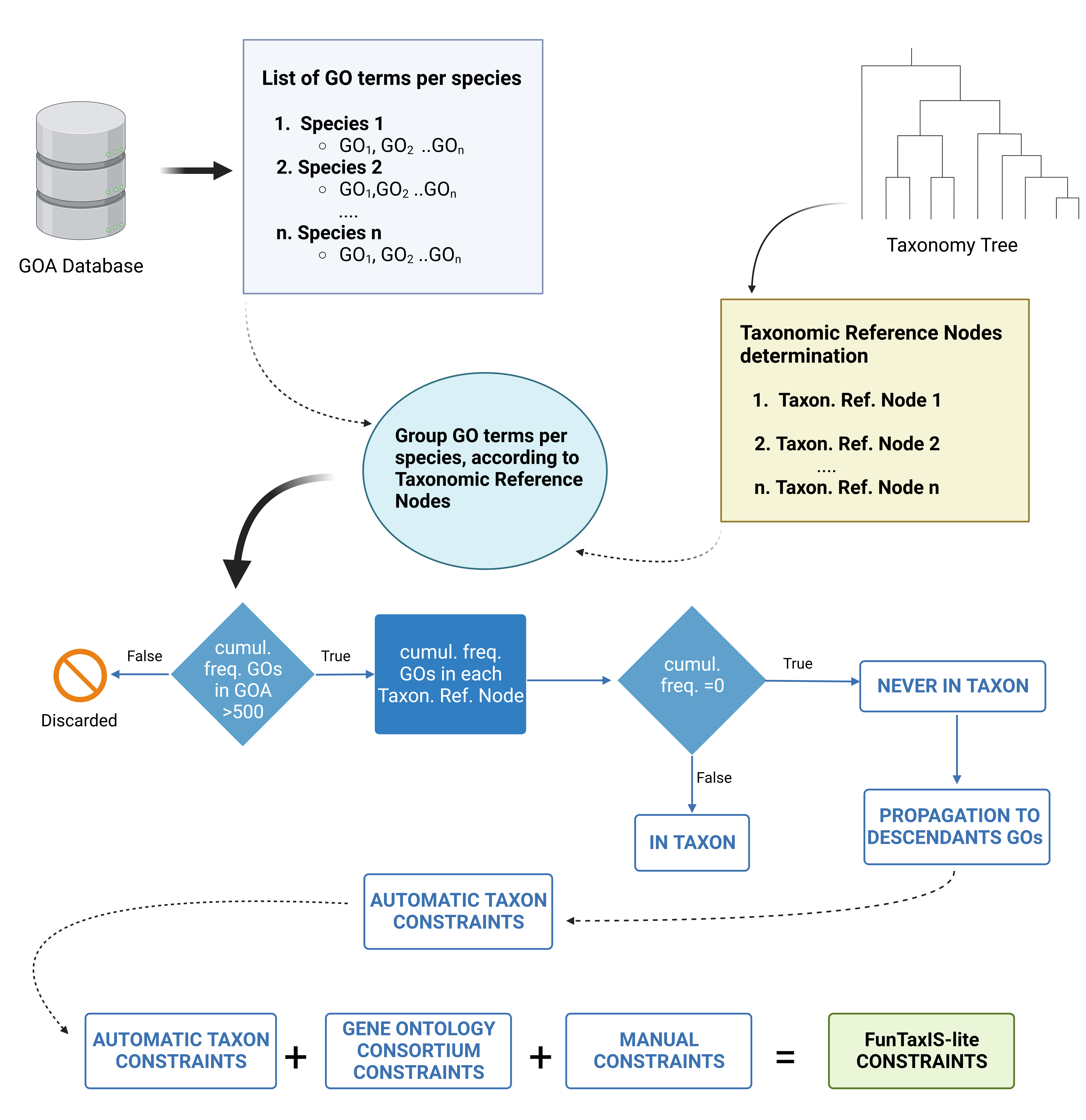
Figure 1a: Schematic representation of the FunTaxIS-lite pipeline. The picture shows the different steps necessary to generate the taxon constraints.
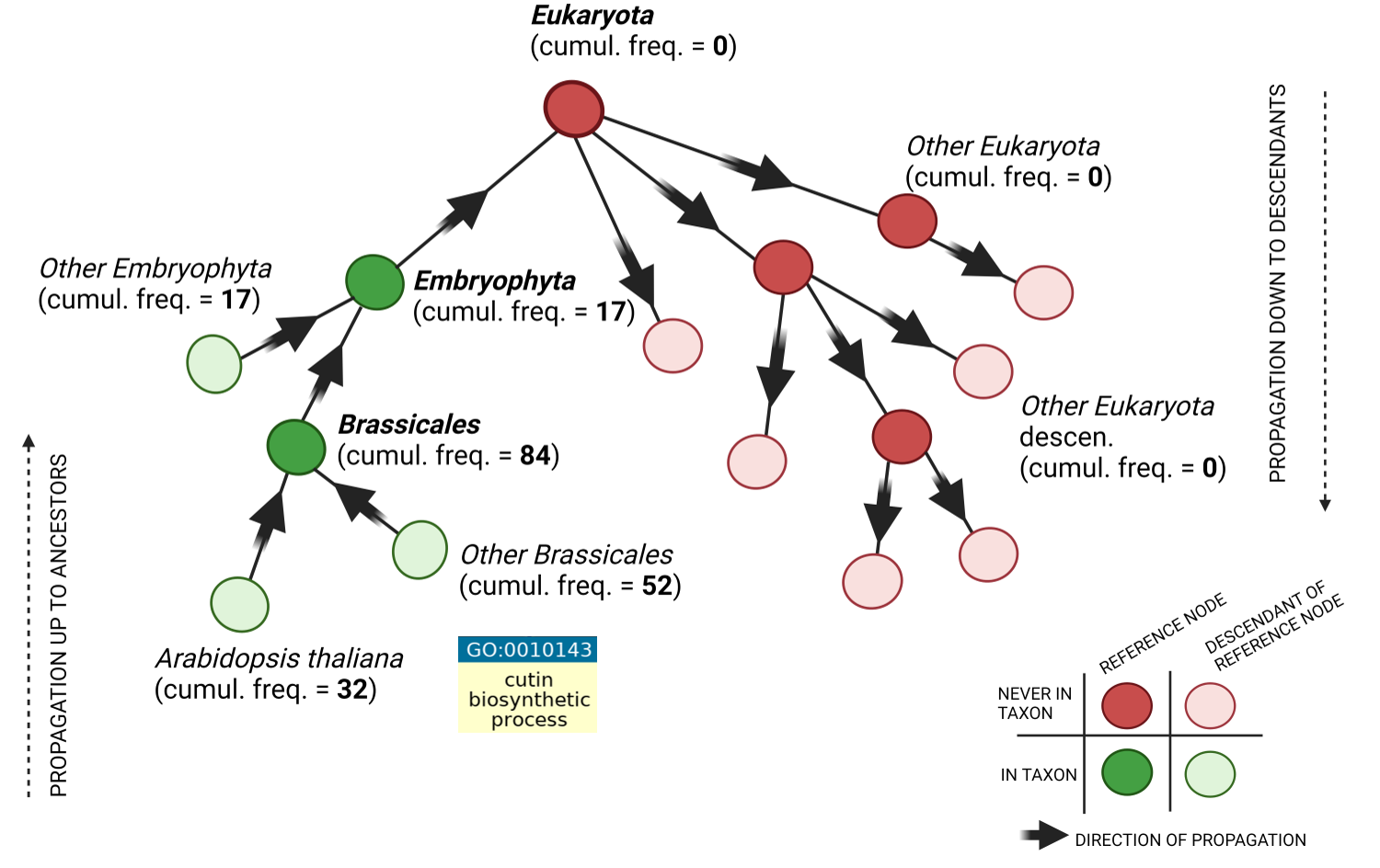
1 GOA database cleaning Figure S1b:The figure shows an example for the GO term GO:0010143 “cutin biosynthetic process”. This term is allowed for plant species since its cumulative frequency is above 0
for both Brassicales and Embryophyta, while it is forbidden for other Eukaryotes since the frequency is 0 for Eukaryota reference node. The figure also shows that annotations coming from
the model organism Arabidopsis thaliana contribute to its “closest” reference node (Brassicales) whilst they do not contribute to the upper node Embryophyta that instead receives annotations
rom other organisms.
The first step requires a cleaning process of the raw Gene Ontology Annotation (GOA), provided as a GO Annotated File (GAF) format by the Gene Ontology Consortium (GOC).
The cleaning step is required to remove either uninformative or peculiar data. For instance, annotations with the curator evidence code “ND” annotations (No biological Data available),
with “NOT” qualifier (protein does NOT perform a specific function), root GO terms (GO:0005575, GO:0008150 and GO:0003674), and annotations with tag “RNAcentral” and “environmental samples”
are removed from the database.
2 Taxonomic Reference Nodes determination
A pivotal step in the FunTaxIS-lite pipeline is to determine a list of taxonomic reference nodes to be used to group organisms and their respective annotation contribution,
that share common biological features. For each reference taxonomic node, a list of taxonomic constraints is then produced and inherited by all organisms subsumed by that reference taxonomic node.
Our approach to identifying reference taxonomic nodes for functional constraints involves balancing two objectives:
1) having a reliable set of constraints for each species subsumed by the corresponding reference taxon;
2) covering a broad range of the taxonomy hierarchy.
This resulted in a total of 240 reference taxonomic nodes representative of all the metabolic/signaling pathways performed by the organisms they subsume.
For example, it is crucial to have a solid list of disallowed functions (e.g. photosynthesis for animals or nervous system for plants). However, due to the inhomogeneity of distribution
of annotations in the GOA, the annotation coverage varies widely among taxonomic ranks, leading to two categories of reference taxonomic nodes:
1) "Reliable taxonomic nodes" are highly representative nodes in the taxonomy hierarchy that group well-annotated branches thanks to the presence of model organisms that are extensively
studied and rich in functional features.
2) "Unreliable taxonomic nodes" are generic nodes in the taxonomy hierarchy with the purpose of grouping poorly annotated branches with limited available knowledge to generate a strong
set of constraints.
3 Grouping GOs and cumulative frequencies calculation
The annotations present in the GOA database are grouped by organisms and for each of them the list of all the associated GO terms is produced, including details about the frequency,
evidence code, and ontology. All species and their annotations are traced back to their “closest” reference parent node. Subsequently, for each GO term of the reference node,
the cumulative frequency over its descendants is calculated.
4 Creation of “never in” GO Taxon Constraints
Once the cumulative frequency of each GO term is obtained for every reference taxon node, the “never-in” constraints are generated following two main steps:
● Cut-off 500 : Only GO terms with a cumulative frequency in the whole GOA that is >= 500 are considered. The cut-off of 500 has been chosen after different tests on a range of
thresholds (see Supplement 3) and guarantees that a certain GO term is relatively spread in the GOA and can be reasonably considered as a candidate for which a constraint can be defined for
certain species. Finally, a GO term for which the cumulative frequency in a reference node is 0 is tagged as “never-in” for that reference node.
Figure S1b provides an illustration of how the cumulative frequency of each GO term for each species contributes to its taxonomic reference node.
● Propagation : the “never-in” generated in the previous step are then propagated down the GO graph following the “true path rule” that governs the graph, i.e. if a parent node is
“false” then its child nodes inherit the ”false” property. As a result, GO terms that may have been discarded by the aforementioned cut-off of 500 could be recovered as “never-in” for
that taxonomic reference node (Figure S1c).
Figure S1c: Propagation of “never-in” over the GO graph. The "true path rule" that governs the GO graph dictates that properties such as "never-in" constraints are passed down through
the graph. In the figure, cyan and light blue nodes report terms that are “in-taxon” while red ones indicate “never-in-taxon” terms. In particular, light red nodes with a cumulative frequency
in GOA < 500 inherit the property from red nodes that have a cumulative frequency > 500. As a result, many GO terms that were not initially considered are now recovered and then considered for
the creation of taxonomic constraints. 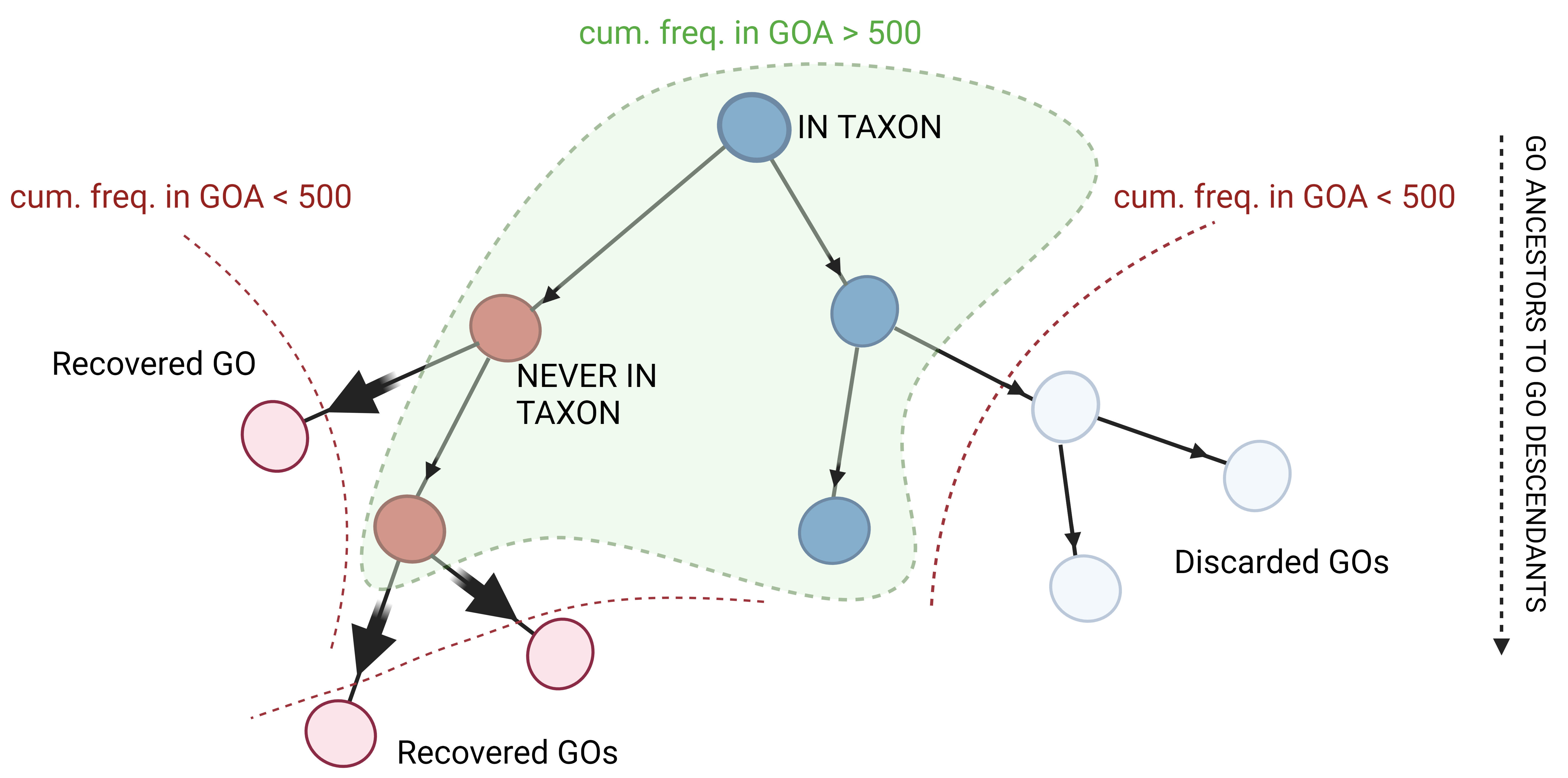
5 Merging automatic, consortium and manual constraints
In the final step of FunTaxIS-lite, the automatic constraints generated by the program are combined with those provided by the Gene Ontology Consortium (referred to as "consortium constraints").
The consortium constraints take precedence over the automatic ones and can override them as needed. Unlike the constraints generated by FunTaxIS-lite, which are only "never-in",
the consortium can also provide the "only-in" constraints. "only-in" constraints can contain additional information, as they can be used to annotate only organisms from a specific taxon,
which automatically makes these GO terms "never-in" for all other species in the taxonomy tree. As a result, the "only-in" constraints are converted into "never-in" and added to the existing
constraints generated by FunTaxIS-lite to increase the total number of constraints. Additionally, to correct errors caused by incorrect annotations in the GOA, a brief list of
"manual constraints" is created based on first-hand observation of annotation issues. These constraints are given the highest priority. In summary, the tool assigns priorities to the different
types of constraints as follows: "manual constraints" have the highest priority, followed by "consortium constraints", and finally, "automatic constraints" have the lowest priority.
Taxonomic Reference Nodes
All the 171 Taxonomic Reference Nodes are reported in the table below."Reliable taxonomic nodes" are highly representative nodes in the taxonomy hierarchy that group well-annotated branches thanks to the presence of model organisms that are extensively studied and rich in functional features. "Unreliable taxonomic nodes" are generic nodes in the taxonomy hierarchy with the purpose of grouping poorly annotated branches with limited available knowledge to generate a strong set of constraints.
Efficacy of the taxonomic constraints. Some examples
The output generated by FunTaxIS-lite is useful for both curators and biologists i) to investigate specific functions oddly absent in some taxa, ii) to spot and then remove possible errors
in the database, and iii) to refine the output of automatic protein function prediction tools. For the latter, FunTaxiS-lite has been assessed using
Argot2.5 (Lavezzo et al.), our in-house tool for automated protein function prediction.
We chose 4 different species (Latimeria chalumnae, Amborella trichopoda, Pseudomonas fluorescens SBW25 and Saccharomyces kudriavzevii IFO 1802) and performed whole-proteome annotation
for each of them by simulating knowledge loss for these organisms. The GO terms predicted have been filtered out using FunTaxIS-lite constraints.
For Latimeria chalumnae (NCBI taxid 7897), 5314 “never-in” constraints were generated by FunTaxIS-lite, and during the prediction 452 of them were found in Argot2.5’s result, hence were filtered out. Some examples:
- GO:0006696, ergosterol biosynthetic process
- GO:0044823, retroviral integrase activity
- GO:0033072, vancomycin biosynthetic process
- GO:0039654, fusion of virus membrane with host endosome membrane
- GO:0009011, starch synthase activity
- GO:1900231, regulation of single-species biofilm formation on inanimate substrate
- GO:0039693, viral DNA genome replication
- GO:0009372, quorum sensing
- GO:0030115, S-layer
- GO:0030153, bacteriocin immunity
- GO:0009061, anaerobic respiration
For Amborella trichopoda (NCBI taxid 13333), 22279 “never-in” constraints were generated by FunTaxIS-lite, and during the prediction 887 of them were found in Argot2.5’s result, hence were filtered out. Some examples:
- GO:0030573, bile acid catabolic process
- GO:0006695, cholesterol biosynthetic process
- GO:0042953, lipoprotein transport
- GO:0009246, enterobacterial common antigen biosynthetic process
- GO:0015948, methanogenesis
- GO:0030283, testosterone dehydrogenase [NAD(P)] activity
- GO:0044010, single-species biofilm formation
- GO:0019062, virion attachment to host cell
- GO:0019083, viral transcription
- GO:0005902, microvillus
For Pseudomonas fluorescens SBW25 (NCBI taxid 216595), 26917 “never-in” constraints were generated by FunTaxIS-lite, and during the prediction 3170 of them were found in Argot2.5’s result, hence were filtered out. Some examples:
- GO:0042383, sarcolemma
- GO:0001772, immunological synapse
- GO:0042476, odontogenesis
- GO:0060250, germ-line stem-cell niche homeostasis
- GO:0050873, brown fat cell differentiation
- GO:0043005, neuron projection
- GO:0009887, animal organ morphogenesis
- GO:0030220, platelet formation
- GO:0070997, neuron death
- GO:0035994, response to muscle stretch
- GO:0060325, face morphogenesis
- GO:0043588, skin development
- GO:0030097, hemopoiesis
For Saccharomyces kudriavzevii IFO 1802 (NCBI taxid 226230), 22009 “never-in” constraints were generated by FunTaxIS-lite, and during the prediction 389 of them were found in Argot2.5’s result, hence were filtered out. Some examples:
- GO:0015979, photosynthesis
- GO:0001568, blood vessel development
- GO:0009579, thylakoid
- GO:0006703, estrogen biosynthetic process
- GO:0042953, lipoprotein transport
- GO:0009740, gibberellic acid mediated signaling pathway
- GO:0004687, myosin light chain kinase activity
- GO:0047696, beta-adrenergic receptor kinase
- GO:0106277, biliverdin reductase (NADP+) activity
- GO:0009736, cytokinin-activated signaling pathway
Last updates
All the taxonomic constraints have been generated starting from the latest available releases of GO, GOA and Taxonomy Tree (Last update: 2024-01-19 11:40:20).


