Menu
G4 virus
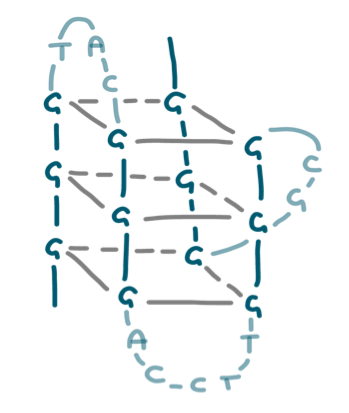
G-quadruplexes are non-canonical nucleic acid structures that control transcription, replication, recombination in organisms. G-quadruplexes are present in eukaryotes, prokaryotes and viruses. In the latter, mounting evidence indicates their key biological activity. Since data on viruses are scattered, we here wished to provide a comprehensive analysis of putative G-quadruplexes in the genome of all known viruses that can infect humans. We show that the presence, distribution and location of G-quadruplexes are features characteristic of each virus class and family. Our statistical analysis proves that their presence within the viral genome is orderly arranged, as indicated by the possibility to correctly assign 67% of viruses to their exact class based on the G-quadruplex classification. For each virus we provide: i) the list of all G-quadruplexes formed by GG-, GGG- and GGGG-islands present in the genome (positive and negative strands), ii) their position in the viral genome along with the known function of that part of the genome, iii) the degree of conservation of each G-quadruplex in its genome context, iv) the statistical significance of G-quadruplex formation. The availability of these data will greatly expedite research on G-quadruplex in viruses, with the possibility to accelerate finding therapeutic opportunities to numerous and fearsome human diseases. Following the link below will get you to the obtained data.
Publication: Lavezzo E, Berselli M, Frasson I, Perrone R, Palù G, Brazzale AR, Richter SN, Toppo S. (2018) G-quadruplex forming sequences in the genome of all known human viruses: A comprehensive guide. PLoS Comput Biol 14(12): e1006675. https://doi.org/10.1371/journal.pcbi.1006675